线性回归之于机器学习,正如Hello World之于编程语言,也如MINST之于深度学习。
首先,我们先定义一些即将用到的数学符号:
| Notations | Meaning | Notations | Meaning |
|---|---|---|---|
| $M$ | Number of parameters $\mathrm w$ | $N$ | Number of instances |
| $\mathrm X={\mathrm x_1,\mathrm x_2,\cdots,\mathrm x_N}^{\mathrm T}$ | $N\times M$ matrix for training | $D$ | Number of features |
| $\mathrm y={y_1,y_2,\cdots,y_N}^\mathrm{T}$ | Set of targets | $y_i$ | Target of instance $i$ |
| $\mathrm{x}_i={x_i^{(1)},x_i^{(2)},\cdots,x_i^{(D)}}^\mathrm{T}$ | Set of features for instance $i$ | $x_i^{(j)}$ | Feature $j$ for instance $i$ |
| $\mathrm w={\omega_1,\omega_2,\cdots,\omega_M}^\mathrm{T}$ | Weights of input $\mathrm x$ | $\omega_i$ | Weight of feature $i$ |
| $\phi={\phi_1,\phi_2,\cdots,\phi_M}^\mathrm{T}$ | Set of functions | $\phi_i(\mathrm x)$ | Function of features |
模型描述
在线性回归中,假设目标值与参数 $\mathrm{w}={\omega_n}$之间线性相关,通过构建损失函数$E$,求解损失函数最小时的参数。也就是说,线性回归试图学习得到如下函数:
公式(1)是线性回归模型的一般形式,看起来不是那么直观。其常见的形式如下:
当$D=1,\phi_j(x)=x^j$时,公式(1)可以表示为:
此时,线性回归就变成了多项式回归。
当$D=M,\phi_j(\mathrm x)=x^{(j)}$时,公式(1)可以表示为:
此时,线性回归就变成了我们通常所说的线性回归—-多元一次方程。当只有一维特征($M=1$) 时,可以得到我们初中就学过的一元一次方程
为使本文通俗易懂,除非作特别说明,本文仿真都以这个一元一次方程为例介绍线性回归
代价函数
线性回归的目的就是使得我们预测得到的$\hat y$与真实值$y$之间的误差最小。这里的误差可以用不同的距离度量,这里我们使用平方和。此时,代价函数就可以表示为
下面我们在二维空间($M=1$)和三维空间($M=2$)画出代价函数图像。这里我们假定$\phi_i(\mathrm x)=x^{(i)}$,$\omega_0,\omega_1,\omega_2$已知,则公式(1)可以分别表示为:
根据公式(6),(7),我们可以得到图1和图2中的直线和二维平面:
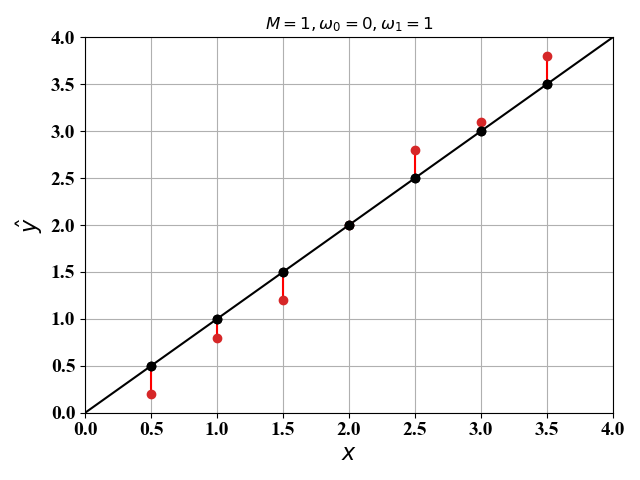
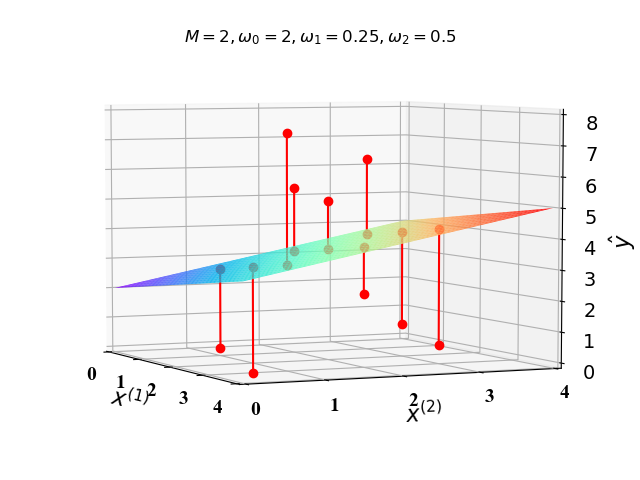
图1和图2中的红色的点是 $\mathrm x$ 对应的真实值 $\mathrm y$ ,红色线段即为误差值。
一个例子
图1和图2展示的是给定参数$\omega_0,\omega_1,\omega_2$下的真实值$y$与预测值$\hat { y}$的误差。不同的参数可以得到不同的误差值,线性回归的目的就是寻找一组参数是的误差最小。下面我们通过图3和图4来说明:
我们假设训练集有3组数据$(x, y)$:$(1, 0.8) (2, 2) (3, 3.1)$ 。我们这里使用一元线性回归,即公式(6),此时线性回归的目的就是找到一条直线$\hat y=\omega_1x+\omega_0$使得这3组数据点离直线最近。
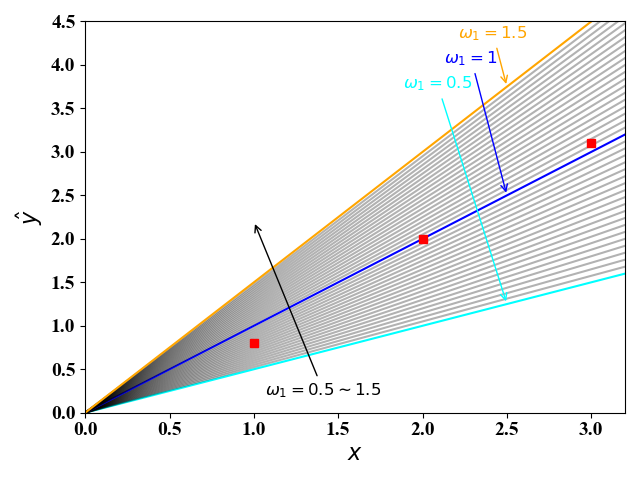
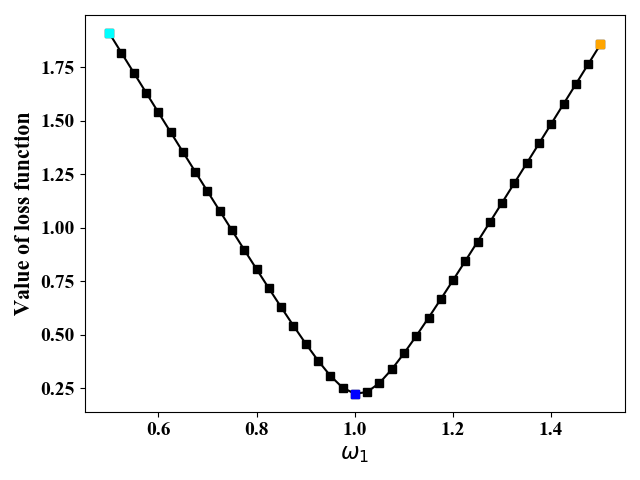
图3画出了当$\omega_0=0,\omega_1=0.5\sim1.5$时,直线$\hat y=\omega_1x+\omega_0$的图像。图4给出了当$\omega_1$取不同值时,代价函数值的变化。从图3和图4可以看出,当$\omega_1=1$时,代价最小。
正规方程与梯度下降
线性回归的本质就是解如下优化问题:
令$\bar{\mathrm w}={\omega_0,\mathrm w},\bar{\phi}={\phi_0,\phi},\phi_0(\mathrm x)=1$,并将问题(8)表示成向量相乘的形式:
公式(9)中,$\bar{\phi}(\mathrm X)$是一个$N\times M+1$维的矩阵:
通过求表达式(8)的Hessian矩阵,可以知道这是一个凸优化问题。那么问题就变得十分简单了,可以用现成的工具来求解:比如CVX, CPLEX, MATLAB等等。这些解法器一般都是通过梯度法(后面会讲解)来求解问题的。当然我们也可以通过凸问题的性质,得到其解析解。
正规方程法
由于误差函数(8)是一个凸函数,所以其导数为0的点就是最优点。为此,我们将$E$对$\mathrm{\bar w}$进行微分求导入下:
由公式(11)可知,给定训练数据$\mathrm X$,我们就可以求出最佳的$\mathrm{\bar w}$。需要注意的是,这里需要求矩阵的逆,计算量比较大,不适合当训练数据较大的情况。这时我们可以通过梯度下降法来求解。
梯度下降法
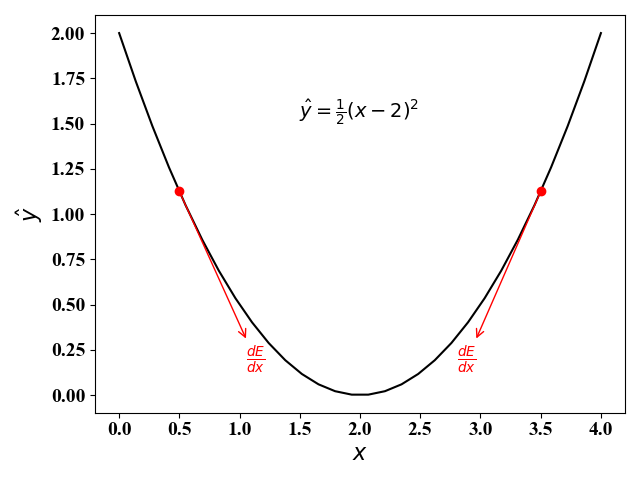
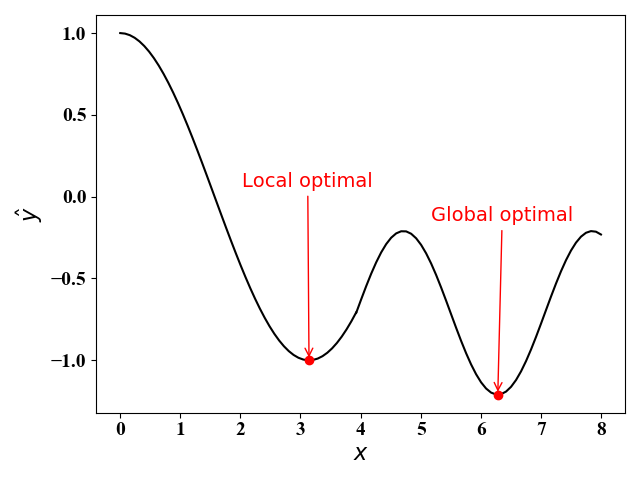
使用梯度下降法,可以对凸问题求得最优解,对非凸问题,可以找到局部最优解。梯度下降法的算法思想如下图5和图6所示:
- 在左图(图5)中,梯度为$\frac{d\hat y}{d x}=x-2$。当$x<2$时,梯度小于零,此时$x$应当向右移动来减小函数值(负梯度方向);当$x>2$时,梯度大于零,此时$x$应当向左移动来减小函数值(负梯度方向)。
- 在右图(图6)中,函数不是凸函数的情况下,使用梯度下降法会得到局部最优解(假定初始值为$x=0$)。当初始值$x=7$时,我们可以得到最优解。因此,初始值对梯度下降法影响较大,我们可以通过随机选择初始值来克服陷入局部最优解的情况。
根据(10)得到的梯度表达式,梯度下降的每一次迭代过程如下:
将公式(13)的矩阵相乘展开可以得到
公式(13)或(14)就是标准的梯度下降法,其中$\eta$是每次迭代的步长大小。
- $\eta$较小时,迭代较慢,当时可以保证收敛到最优解(凸函数的情况下);$\eta$较大时,函数值下降较快,但容易发生震荡。
- 每次迭代时,需要使用所有的样本点$\mathrm x_i,i=1,2,\cdots,N$。当数据样本点非常大时,开销十分大。
为此,有人提出了随机梯度下降,其迭代公式如下:
随机梯度下降又称连续梯度下降,比较适合于实时系统,即整个数据集$\mathrm x_i$不是可以一次性获得的,但是我们需要作出预测的场景。相较于梯度下降法(14),随机梯度只根据当前样本更新迭代,随机性较大。因此有可能跳出标准梯度下降法的局部最优解。
算法实现
这里我们使用sklearn中波士顿房价的数据集,该数据集有13维特征,506个样例。为简便起见,我们只取前2维特征作为输入($M=D=2,\hat y=\omega_0+\omega_1x^{(1)}+\omega_2x^{(2)}$),前500个作为输入样例,后6个作为预测样例。在算法实现中,我们分别考虑了正规方程法和梯度下降法。并且,考虑到$x^{(1)}$和$x^{(2)}$的取值范围差距较大,我们还考虑了特征值缩放。为此,我们实现了上述四种算法的组合[特征不缩放(特征缩放)+正规方程法(梯度下降法)]。
算法结果
图7给出了上述4种算法的结果:
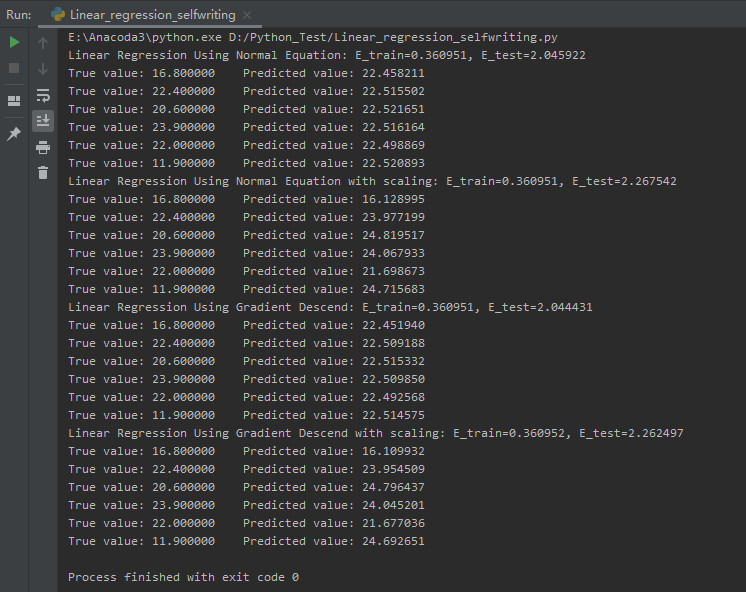
图7中,E_train为训练误差,即前500个样例的真实值与预测值的误差,E_test为预测误差,即最后6个样例的真实值与预测值的误差。由于误差函数对于参数w是凸函数,我们总能得到最优解,即最小的训练误差,所以上述四种方法的训练误差相同。
特征缩放与梯度下降法
图7能得到最小误差函数值,是因为目标函数$E$是参数$\omega_1$和$\omega_2$的凸函数。为方便起见,对于具体实例,我们给出$E$的表达式:
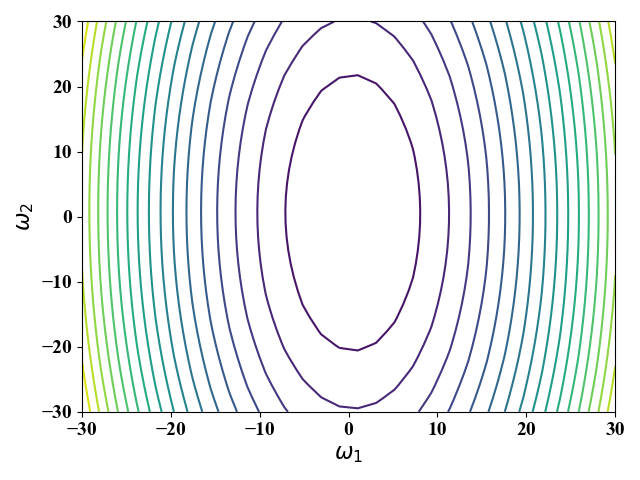
公式(16)中,$\omega_0$与具体样例无关,$\omega_0$的值不改变$E$的图像形状,改变$\omega_0$相当于进行位移,我们这里假定$\omega_0=0$。为此,当给定波士顿房价数据集,即$x^{(1)},x^{(2)},y_i$ 给定时,我们可以画出公式(16)对应的等高线图,图8。
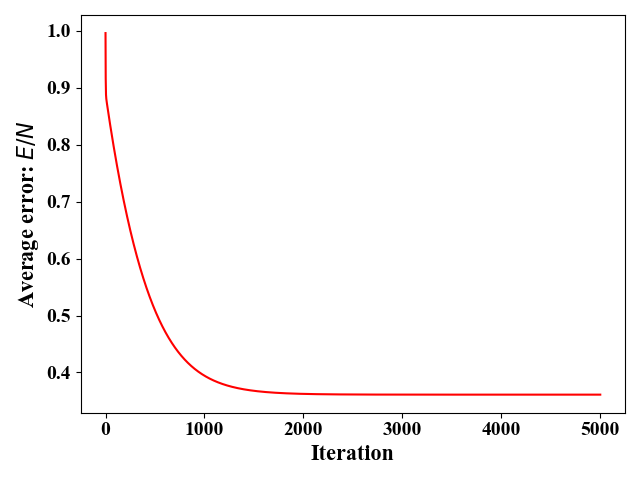
从图8可以看出,当改变$\omega_2$时,$E$变的较快(等高线在$\omega_2$方向较为稀疏)。这是因为$\omega_2$的系数为$x^{(2)}$,而$x^{(2)}$相对于$x^{(1)}$有较大的取值。在这种情况下,对梯度下降法就十分不友好—很容易跳过最优解。也就是说,步长设置要十分小,这就会导致收敛速度慢。在我们这个实例中,步长最大只能设置为$\eta=5e^{-6}$,此时需要差不多30000次迭代才能收敛到最优,如图10所示。
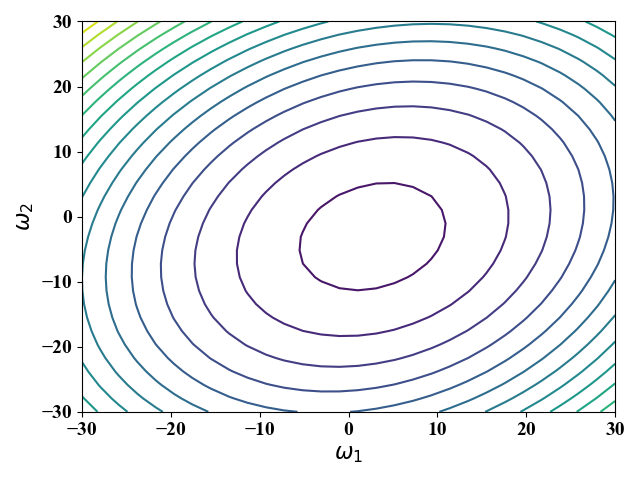
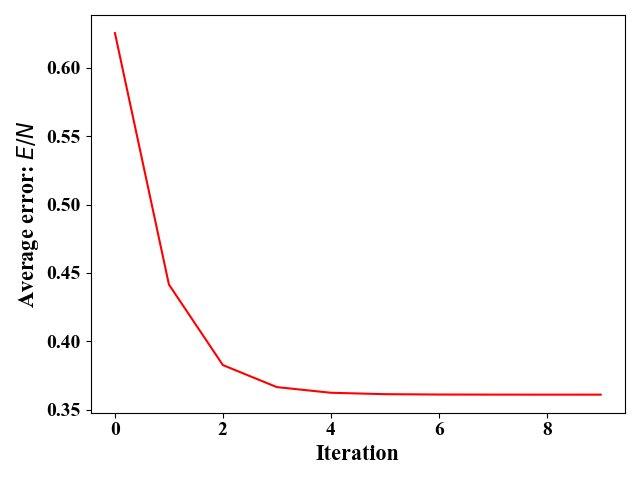
特征缩放是一种解决上述情况下,梯度下降法收敛慢的方法。特征缩放的表达式都十分简单,这里不再赘述,我们这里是直接使用的sklearn库中的preprocessing.StandardScaler()函数对样例进行特征缩放。对$x^{(1)},x^{(2)}$缩放后,我们可以用相同的方式画出对应的等高线图,图9,以及收敛图,图11。经过特征缩放后,图9中等高线在$\omega_1,\omega_2$方向上的稀疏程度差不多。图11中,步长可以设置得较大($\eta=1e^{-3}$),收敛速度变得极快,只需要迭代8次左右就达到最优。
附录
下面给出图1—图11的Python源代码如下:
- 图1和图2的python源代码:
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87
88
89
90import numpy as np
import matplotlib.pyplot as plt
from mpl_toolkits.mplot3d import Axes3D
# Set the format of labels
def LabelFormat(plt):
ax = plt.gca()
plt.tick_params(labelsize=14)
labels = ax.get_xticklabels() + ax.get_yticklabels()
[label.set_fontname('Times New Roman') for label in labels]
font = {'family': 'Times New Roman',
'weight': 'normal',
'size': 16,
}
return font
# 2-d case
omega_0 = 0
omega_1 = 1
data_train = [[0.5, 0.2], [1, 0.8], [1.5, 1.2], [2, 2], [2.5, 2.8], [3, 3.1], [3.5, 3.8]]
x_train = [d[0] for d in data_train]
y_train = [d[1] for d in data_train]
x = np.linspace(0, 4, 30).reshape(30, 1)
y = omega_1 * x + omega_0
x_test = x_train
y_test = y_train
y_hat = omega_1 * x_test
plt.figure()
plt.plot(x, y, 'k-')
for i in range(len(x_test)):
plt.stem([x_test[i], ], [y_test[i], ], linefmt='rx', bottom=y_hat[i], basefmt='ko', markerfmt='C3o',
use_line_collection=True)
# Set the labels
font = LabelFormat(plt)
plt.xlabel('$x$', font)
plt.ylabel('$\hat y$', font)
plt.title('$M=1,\omega_0=0,\omega_1=1$')
plt.xlim(0, 4)
plt.ylim(0, 4)
plt.grid()
plt.show()
# 3-d case
omega_0 = 2
omega_1 = 0.25
omega_2 = 0.5
x1 = np.linspace(0, 4, 30).reshape(30, 1)
x2 = np.linspace(0, 4, 30).reshape(30, 1)
X1, X2 = np.meshgrid(x1, x2)
y_hat = omega_0 + omega_1 * X1 + omega_2 * X2
fig = plt.figure()
ax = fig.gca(projection='3d')
x1_test=np.array([1,2,3])
x2_test=np.array([1,2,3])
X1_test, X2_test = np.meshgrid(x1_test, x2_test)
y_test = omega_0 + omega_1 * X1_test + omega_2 * X2_test+8*np.random.rand(3,3)-4
ax.plot_surface(X1, X2, y_hat, cmap='rainbow')
for i in range(len(x1_test)):
for j in range(len(x2_test)):
y_predict= omega_0 + omega_1 * x1_test[i] + omega_2 * x2_test[j]
ax.plot([x1_test[i],x1_test[i]],[x2_test[j],x2_test[j]],[y_test[i][j],y_predict],'r-o')
# Set the labels
font = LabelFormat(plt)
ax.set_xlabel('$x^{(1)}$', font)
ax.set_ylabel('$x^{(2)}$', font)
ax.set_zlabel('$\hat y$', font)
ax.set_xlim(0, 4)
ax.set_ylim(0, 4)
ax.set_zlim(0, 8)
ax.set_xticks([0,1,2,3,4])
ax.set_yticks([0,1,2,3,4])
ax.set_title('$M=2,\omega_0=2,\omega_1=0.25,\omega_2=0.5$')
# Customize the view angle so it's easier to see that the scatter points lie
ax.view_init(elev=5., azim=-25)
plt.show()
- 图3和图4的python源代码:
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87
88
89
90
91
92
93
94
95
96
97
98
99
100
101import numpy as np
import matplotlib.pyplot as plt
from matplotlib.colors import ListedColormap
import matplotlib as mpl
import math
# Set the format of labels
def LabelFormat(plt):
ax = plt.gca()
plt.tick_params(labelsize=14)
labels = ax.get_xticklabels() + ax.get_yticklabels()
[label.set_fontname('Times New Roman') for label in labels]
font = {'family': 'Times New Roman',
'weight': 'normal',
'size': 16,
}
return font
# Plot the training points: different
def PlotTrainPoint(X):
for i in range(0, len(X)):
plt.plot(X[i][0], X[i][1], 'rs', markersize=6, markerfacecolor="r")
# Loss function--Square Error function
def LossFunction(Y, predictedY):
lengthY = len(Y)
error = 0
for i in range(lengthY):
error += pow(Y[i] - predictedY[i], 2)
return math.sqrt(error)
trainData = [[1, 0.8], [2, 2], [3, 3.1]]
# Predicted function: y=\omega_1*x+\omega_0 Here \omega_0 is assumed to be 0 for simplifcity
x = np.linspace(0, 4, 30).reshape(30, 1)
omega_1 = np.linspace(0.5, 1.5, 41).reshape(41, 1)
omega_0 = 0
y_hat = []
#Get the value of x and y in the trainData
x_train = [d[0] for d in trainData]
y_train = [d[1] for d in trainData]
error_all = []
# Plot the figure to show the function: y=\omega_1*x+\omega_0
for i in range(len(omega_1)):
y_hat.append(omega_1[i] * x)
if omega_1[i]==0.5:
plt.plot(x, y_hat[i], color='cyan', alpha=1)
elif omega_1[i]==1:
plt.plot(x, y_hat[i], color='blue', alpha=1)
elif omega_1[i]==1.5:
plt.plot(x, y_hat[i], color='orange', alpha=1)
else:
plt.plot(x, y_hat[i], color='black', alpha=0.3)
# Compute the errors for each omega_1
error_all.append(LossFunction(y_train, omega_1[i].T*x_train+omega_0))
# Set the axis
font=LabelFormat(plt)
PlotTrainPoint(trainData)
# Label the critical points
plt.annotate('$\omega_1=1.5$', xy=(2.5, 2.5*1.5), xycoords='data',
xytext=(-35, 35), textcoords='offset points', color='orange', fontsize=12, arrowprops=dict(arrowstyle="->",
connectionstyle="arc,rad=90", color='orange'))
plt.annotate('$\omega_1=1$', xy=(2.5, 2.5*1), xycoords='data',
xytext=(-45, 95), textcoords='offset points', color='b', fontsize=12, arrowprops=dict(arrowstyle="->",
connectionstyle="arc,rad=90", color='b'))
plt.annotate('$\omega_1=0.5$', xy=(2.5, 2.5*0.5), xycoords='data',
xytext=(-75, 155), textcoords='offset points', color='cyan', fontsize=12, arrowprops=dict(arrowstyle="->",
connectionstyle="arc,rad=90", color='cyan'))
plt.annotate('$\omega_1=0.5\sim 1.5$', xy=(1, 2.2), xycoords='data',
xytext=(8, -125), textcoords='offset points', color='k', fontsize=12, arrowprops=dict(arrowstyle="->",
connectionstyle="arc,rad=90", color='k'))
plt.xlabel('$x$',font)
plt.ylabel('$\hat y$',font)
plt.xlim([0,3.2])
plt.ylim([0,4.5])
plt.show()
# Show the error when omega_1 changes
plt.figure()
font=LabelFormat(plt)
plt.plot(omega_1,error_all, 'k-s')
error_min=min(error_all)
index_min=error_all.index(error_min)
print(index_min)
# plot the error at the given three point
plt.plot(omega_1[index_min],error_min,'bs')
plt.plot(omega_1[0],error_all[0],'cyan',marker='s')
plt.plot(omega_1[-1],error_all[-1],'orange',marker='s')
plt.xlabel('$\omega_1$', font)
plt.ylabel('Value of loss function', font)
plt.show() - 图5和图6的python源代码:
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64import numpy as np
import matplotlib.pyplot as plt
# Set the format of labels
def LabelFormat(plt):
ax = plt.gca()
plt.tick_params(labelsize=14)
labels = ax.get_xticklabels() + ax.get_yticklabels()
[label.set_fontname('Times New Roman') for label in labels]
font = {'family': 'Times New Roman',
'weight': 'normal',
'size': 16,
}
return font
x = np.linspace(0, 4, 30).reshape(30, 1)
y=(x-2)**2/2
plt.figure()
plt.plot(x,y,'k-')
plt.plot(3.5,1.5**2/2,'ro')
plt.annotate('$\\frac{dE}{dx}$', xy=(3.5, 1.5**2/2), xycoords='data',
xytext=(-60, -125), textcoords='offset points',color='r', fontsize=14, arrowprops=dict(arrowstyle="<-",
connectionstyle="arc,rad=90", color='r'))
plt.plot(0.5,1.5**2/2,'ro')
plt.annotate('$\\frac{dE}{dx}$', xy=(0.5, 1.5**2/2), xycoords='data',
xytext=(48, -125), textcoords='offset points',color='r', fontsize=14, arrowprops=dict(arrowstyle="<-",
connectionstyle="arc,rad=90", color='r'))
plt.annotate('$\hat y=\\frac{1}{2}(x-2)^2$', xy=(0.25, 1.75**2/2), xycoords='data',
xytext=(108, 0), textcoords='offset points',color='k', fontsize=14, arrowprops=dict(arrowstyle="<-",
connectionstyle="arc,rad=90", color='w'))
# Set the labels
font = LabelFormat(plt)
plt.xlabel('$x$', font)
plt.ylabel('$\hat y$', font)
plt.show()
# To plot figure 6
x1 = np.linspace(0, 5/4.0*np.pi, 50).reshape(50, 1)
y1=np.cos(x1)
x2 = np.linspace(5/4.0*np.pi, 8, 50).reshape(50, 1)
y2=0.5*np.cos(2*x2+1*np.pi)-0.71
plt.figure()
plt.plot(x1,y1,'k-')
plt.plot(x2,y2,'k-')
plt.plot(np.pi,-1,'ro')
plt.annotate('Local optimal', xy=(np.pi, -1), xycoords='data',
xytext=(-48, 125), textcoords='offset points',color='r', fontsize=14, arrowprops=dict(arrowstyle="->",
connectionstyle="arc,rad=90", color='r'))
plt.plot(np.pi*2,-1.21,'ro')
plt.annotate('Global optimal', xy=(2*np.pi, -1.21), xycoords='data',
xytext=(-48, 125), textcoords='offset points',color='r', fontsize=14, arrowprops=dict(arrowstyle="->",
connectionstyle="arc,rad=90", color='r'))
# Set the labels
font = LabelFormat(plt)
plt.xlabel('$x$', font)
plt.ylabel('$\hat y$', font)
plt.show() - 图7和图11的python源代码:
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87
88
89
90
91
92
93
94
95
96
97
98
99
100
101
102
103
104
105
106
107
108
109
110
111
112
113
114
115
116
117
118
119
120
121
122
123
124
125
126
127
128
129
130
131
132
133
134
135
136
137
138
139
140
141
142
143
144
145
146
147
148
149
150
151
152
153
154
155
156
157
158
159
160
161
162
163
164
165
166
167
168
169
170
171
172
173
174
175
176
177
178
179
180
181
182
183
184
185
186
187
188
189
190
191
192
193
194
195
196
197
198
199
200
201
202
203
204
205
206
207
208
209
210
211
212
213
214
215
216
217
218
219
220
221
222
223
224
225
226
227
228
229
230
231
232
233
234
235
236
237
238
239
240
241
242
243
244
245
246
247
248
249
250
251
252
253
254
255
256
257
258
259
260
261
262
263
264
265
266
267
268
269
270
271
272
273
274
275
276
277
278
279
280
281
282
283# -*- coding: utf-8 -*-
# @Time : 2020/4/7 11:28
# @Author : tengweitw
import numpy as np
from sklearn.datasets import load_boston
import matplotlib.pyplot as plt
from mpl_toolkits.mplot3d import Axes3D
from sklearn import preprocessing
def Linear_regression_normal_equation(train_data, train_target, test_data, test_target):
# the 1st column is 1 i.e., x_0=1
temp = np.ones([np.size(train_data, 0), 1])
# X is a 500*(1+2)-dim matrix
X = np.concatenate((temp, train_data), axis=1)
# Normal equation
w_bar = np.matmul(np.linalg.inv(np.matmul(X.T, X)), np.matmul(X.T, train_target))
# Training Error
y_predict_train = np.matmul(X, w_bar)
E_train = np.linalg.norm(y_predict_train - train_target)/len(y_predict_train)
# Predicting
x0 = np.ones((np.size(test_data, 0), 1))
test_data1 = np.concatenate((x0, test_data), axis=1)
y_predict_test = np.matmul(test_data1, w_bar)
# Prediction Error
E_test = np.linalg.norm(y_predict_test - test_target)/len(y_predict_test)
return y_predict_test, E_train, E_test
def Linear_regression_normal_equation_scale(train_data, train_target, test_data, test_target):
# Data processing: scaling
# For training data
ss = preprocessing.StandardScaler()
ss.partial_fit(train_data)
train_data_scale = ss.fit_transform(train_data)
# For testing data
ss.partial_fit(test_data)
test_data_scale = ss.fit_transform(test_data)
# the 1st column is 1 i.e., x_0=1
temp = np.ones([np.size(train_data_scale, 0), 1])
# X is a 500*(1+2)-dim matrix
X = np.concatenate((temp, train_data_scale), axis=1)
# Normal equation
w_bar = np.matmul(np.linalg.inv(np.matmul(X.T, X)), np.matmul(X.T, train_target))
# Training Error
y_predict_train = np.matmul(X, w_bar)
E_train = np.linalg.norm(y_predict_train - train_target) / len(y_predict_train)
# Predicting
x0 = np.ones((np.size(test_data_scale, 0), 1))
test_data1 = np.concatenate((x0, test_data_scale), axis=1)
y_predict_test = np.matmul(test_data1, w_bar)
# Prediction Error
E_test = np.linalg.norm(y_predict_test - test_target) / len(y_predict_test)
return y_predict_test, E_train, E_test
def Linear_regression_gradient_descend(train_data, train_target, test_data, test_target):
# learning rate
eta = 5e-6
M = np.size(train_data, 1)
N = np.size(train_data, 0)
w_bar = np.zeros((M + 1, 1))
# the 1st column is 1 i.e., x_0=1
temp = np.ones([N, 1])
# X is a N*(1+M)-dim matrix
X = np.concatenate((temp, train_data), axis=1)
train_target = np.mat(train_target).T
iter = 0
num_iter = 5000
E_train = np.zeros((num_iter, 1))
while iter < num_iter:
temp = np.matmul(X, w_bar) - train_target
w_bar = w_bar - eta * np.matmul(X.T, temp)
# Predicting training data
y_predict_train = np.matmul(X, w_bar)
# Training Error
E_train[iter]=np.linalg.norm(y_predict_train - train_target)/len(y_predict_train)
iter += 1
# Predicting
x0 = np.ones((np.size(test_data, 0), 1))
test_data1 = np.concatenate((x0, test_data), axis=1)
y_predict_test = np.matmul(test_data1, w_bar)
# Prediction Error
E_test = np.linalg.norm(y_predict_test.ravel()- test_target)/len(y_predict_test)
return y_predict_test, E_train, E_test
def Linear_regression_gradient_descend_scale(train_data, train_target, test_data, test_target):
# Data processing: scaling
# For training data
ss = preprocessing.StandardScaler()
ss.partial_fit(train_data)
train_data_scale = ss.fit_transform(train_data)
# For testing data
ss.partial_fit(test_data)
test_data_scale = ss.fit_transform(test_data)
# learning rate
eta = 1e-3
M = np.size(train_data_scale, 1)
N = np.size(train_data_scale, 0)
w_bar = np.zeros((M + 1, 1))
# the 1st column is 1 i.e., x_0=1
temp = np.ones([N, 1])
# X is a N*(1+M)-dim matrix
X = np.concatenate((temp, train_data_scale), axis=1)
train_target = np.mat(train_target).T
iter = 0
num_iter = 10
E_train = np.zeros((num_iter, 1))
while iter < num_iter:
temp = np.matmul(X, w_bar) - train_target
w_bar = w_bar - eta * np.matmul(X.T, temp)
# Predicting training data
y_predict_train = np.matmul(X, w_bar)
# Training Error
E_train[iter]=np.linalg.norm(y_predict_train - train_target)/len(y_predict_train)
iter += 1
# Predicting
x0 = np.ones((np.size(test_data_scale, 0), 1))
test_data1 = np.concatenate((x0, test_data_scale), axis=1)
y_predict_test = np.matmul(test_data1, w_bar)
# Prediction Error
E_test = np.linalg.norm(y_predict_test.ravel()- test_target)/len(y_predict_test)
return y_predict_test, E_train, E_test
# Set the format of labels
def LabelFormat(plt):
ax = plt.gca()
plt.tick_params(labelsize=14)
labels = ax.get_xticklabels() + ax.get_yticklabels()
[label.set_fontname('Times New Roman') for label in labels]
font = {'family': 'Times New Roman',
'weight': 'normal',
'size': 16,
}
return font
def Plot_error_vs_omega(train_data,train_target):
# ---------Show the contour of E with respect to omegas---------------------
x1 = train_data[:, 0]
x2 = train_data[:, 1]
omega_1 = np.linspace(-30, 30, 30)
omega_2 = np.linspace(-30, 30, 30)
Y_hat = np.zeros((len(omega_1),len( omega_2)))
for i in range(len(omega_1)):
for j in range(len(omega_2)):
for k in range(len(train_data)):
temp=train_target[k] - (omega_1[i] * x1[k] + omega_2[j] * x2[k])
Y_hat[i][j] = Y_hat[i][j] + np.square(temp)
fig = plt.figure()
plt.contour(omega_2,omega_1,Y_hat,20)
# Set the labels
font = LabelFormat(plt)
plt.xlabel('$\omega_1$', font)
plt.ylabel('$\omega_2$', font)
plt.show()
def Plot_error_vs_omega_scale(train_data, train_target):
# ---------Show the contour of E with respect to omegas---------------------
# Data processing: scaling
# For training data
ss = preprocessing.StandardScaler()
ss.partial_fit(train_data)
train_data_scale = ss.fit_transform(train_data)
x1 = train_data_scale[:, 0]
x2 = train_data_scale[:, 1]
omega_1 = np.linspace(-30, 30, 30)
omega_2 = np.linspace(-30, 30, 30)
Y_hat = np.zeros((len(omega_1), len(omega_2)))
for i in range(len(omega_1)):
for j in range(len(omega_2)):
for k in range(len(train_data_scale)):
temp = train_target[k] - (omega_1[i] * x1[k] + omega_2[j] * x2[k])
Y_hat[i][j] = Y_hat[i][j] + np.square(temp)
fig = plt.figure()
plt.contour(omega_2, omega_1, Y_hat, 20)
# Set the labels
font = LabelFormat(plt)
plt.xlabel('$\omega_1$', font)
plt.ylabel('$\omega_2$', font)
plt.show()
if __name__ == '__main__':
# load house price of Boston
data, target = load_boston(return_X_y=True)
# The number of selected features
M = 2
# The first 500 data for training
train_data = data[0:500, 0:0 + M]
train_target = target[0:500]
train_target.reshape(len(train_data), 1)
# ------------------------------
# The last 6 data for testing
test_data = data[500:, 0:0 + M]
test_target = target[500:]
# To show the contour of error function E with respect to omega
# We can see that it's a convex function, not easy for gradient descend
Plot_error_vs_omega(train_data, train_target)
Plot_error_vs_omega_scale(train_data, train_target)
#---------------------------------#
y_predict_normal_equation, E_train,E_test = Linear_regression_normal_equation(train_data, train_target, test_data,
test_target)
print("Linear Regression Using Normal Equation: E_train=%f, E_test=%f" % (E_train,E_test))
for i in range(len(test_data)):
print("True value: %f Predicted value: %f" % (test_target[i], y_predict_normal_equation[i]))
# ---------------------------------#
y_predict_normal_equation_scale, E_train,E_test = Linear_regression_normal_equation_scale(train_data, train_target,
test_data, test_target)
print("Linear Regression Using Normal Equation with scaling: E_train=%f, E_test=%f" % (E_train,E_test))
for i in range(len(test_data)):
print("True value: %f Predicted value: %f" % (test_target[i], y_predict_normal_equation_scale[i]))
# ---------------------------------#
y_predict_gradient_descent, E_train,E_test = Linear_regression_gradient_descend(train_data, train_target, test_data,
test_target)
print("Linear Regression Using Gradient Descend: E_train=%f, E_test=%f" % (E_train[-1],E_test))
for i in range(len(test_data)):
print("True value: %f Predicted value: %f" % (test_target[i], y_predict_gradient_descent[i]))
plt.figure()
plt.plot(E_train,'r-')
# Set the labels
font = LabelFormat(plt)
plt.xlabel('Iteration', font)
plt.ylabel('Average error: $E/N$', font)
plt.show()
# ---------------------------------#
y_predict_gradient_descent_scale, E_train,E_test = Linear_regression_gradient_descend_scale(train_data, train_target,
test_data, test_target)
print("Linear Regression Using Gradient Descend with scaling: E_train=%f, E_test=%f" % (E_train[-1],E_test))
for i in range(len(test_data)):
print("True value: %f Predicted value: %f" % (test_target[i], y_predict_gradient_descent_scale[i]))
plt.figure()
plt.plot(E_train,'r-')
# Set the labels
font = LabelFormat(plt)
plt.xlabel('Iteration', font)
plt.ylabel('Average error: $E/N$', font)
plt.show()

